More about prepare-rpbp-genome#
As part of the index creation step, Rp-Bp extract all putative ORFs and assign to each one a label based on its position relative to the annotated transcript-exon structure. Labels can be useful to quickly identify certain ORF types that may be of particular interest, e.g. upstream ORFs, or ORFs from non-coding RNAs.
Note
The ORF “id” is of the form transcript_seqname:start-end:strand. seqname is the chromosome or contig. The start codon is included, but the stop codon is not. The host transcript “id” should not contain underscores!
Hint
In some cases, the ORF label may not be consistent with the host transcript, as reported by the ORF “id”. To resolve such seemingly incoherent assignments, compatible transcripts are reported for each ORF in <genome_name>.orfs-labels.annotated[.orf_note].tab.gz and shown in the prediction dashboard (see Visualization and quality control).
Categories of Ribo-seq ORFs#
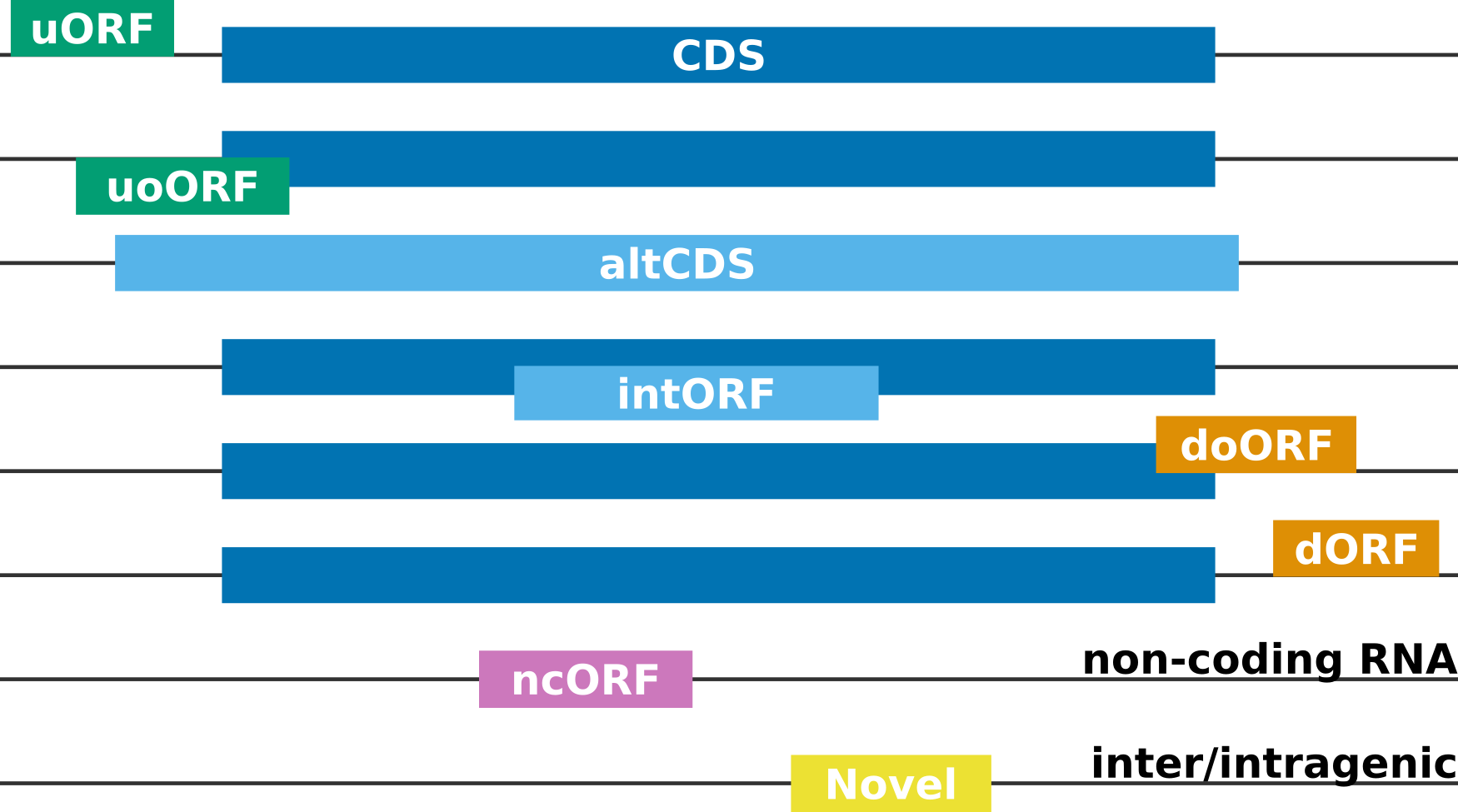
CDS: Canonical (annotated) coding sequence
altCDS: Alternative CDS (e.g. N/C-terminus extension/truncation, alternatively spliced variants, etc.)
intORF: Translation event within a CDS (in- or out-of-frame)
uORF/uoORF: Translation event in the 5’ untranslated region (UTR) of or partially overlapping an annotated protein-coding gene
dORF/doORF: Translation event in the 3’ untranslated region (UTR) of or partially overlapping an annotated protein-coding gene
ncORF: Translation event in an RNA annotated as non-coding (lncRNA, pseudogene, etc.)
Novel: Translation event inter- or intragenic (only when Rp-Bp is run with a de novo assembly, see below)
Labels such as overlap or suspect arise when Rp-Bp is not able to resolve the position of an ORF without ambiguity. In practice, for standard annotations, we do not see these categories.
More about de novo ORF discovery#
For Rp-Bp, there is no difference between annotated and de novo assembled transcripts. In both cases, ORFs are extracted from the transcripts based on the given start and stop codons. However, it is often of scientific interest to identify Novel Ribo-seq ORFs. These are the most interesting, as they do not overlap the annotations at all, but Rp-Bp also identifies Novel altCDS and Novel ncORF.
Hence, when matching RNA-seq is available (same reference genome), we highly recommend to create a de novo assembly. The only requirement is that the assembler produces a valid GTF file (or a format that can be converted to GTF). In a de novo assembly, coding regions are typically not identified (that is what Ribo-seq is for!). However, if your assembly also includes CDS annotations, they must satisfy the start/stop codon GTF2 specifications (stop codon not included in the CDS.)