Visualization and quality control#
Rp-Bp provides two interactive dashboards or web applications, one for read and periodicity quality control, the other to facilitate Ribo-seq ORFs discovery. The latter has an integrated IGV browser for the visual exploration of predicted Ribo-seq ORFs. To navigate the apps is easy, just follow the “hints”. Most items are interactive.
Before you can visualize the results, you need to prepare the input for the dashboards.
How to prepare the input for the dashboards#
Summarizing the profile construction#
To prepare the input for the profile construction dashboard
summarize-rpbp-profile-construction <config> [options]
With the -c/--create-fastqc-reports flag, FastQC reports are be created. For all options, consult the API.
Required input#
Output files from the ORF profile construction step.
Output files#
The base path for these files is: <riboseq_data>/analysis/profile_construction.
<project_name>[.note].read-filtering-counts.csv.gz A CSV file with read counts after each step of the pipeline, one row per sample, with the following columns: “note” (sample name), “raw_data_count” (reads in the original FASTQ), “without_adapters_count” (reads remaining after running Flexbar), “without_rrna_count” (reads remaining after rRNA removal), “genome_count” (genome alignments), “unique_count” (unique genome alignments), “length_count” (periodic reads).
<project_name>[.note].length-distribution.csv.gz A CSV file with counts of aligned reads for each length, one row per sample.
<project_name>[.note][-unique].periodic-offsets.csv.gz A CSV file with all P-site offsets and read lengths for each samples.
<project_name>[.note][-unique].frame-counts.csv.gz A CSV file with P-site adjusted coverage across 3 frames summed across all ORFs for each sample.
Summarizing the Rp-Bp predictions#
To prepare the input for the predictions dashboard
summarize-rpbp-predictions <config> [options]
For all options, consult the API.
Required input#
Output files from the translation prediction step.
Output files#
The base path for these files is: <riboseq_data>/analysis/rpbp_predictions.
<project_name>[.note][-unique][.filtered].predicted-orfs.bed.gz A BED12+ file with the combined predicted translation events from all samples and replicates. Additional columns include sample or replicate, Bayes factor mean and variance, P-site coverage across 3 frames, ORF number, ORF length, label, host transcript biotype, and associated gene id, name and biotype, and compatible transcripts.
<project_name>[.note][-unique][.filtered].igv-orfs.bed.gz A BED12 file with all unique ORFs.
<project_name>.summarize_options.json A json summary file for the app.
<genome_name>.circos_graph_data.json A json file with the ORF distribution across chromosomes.
Hint
Use <project_name>[.note][-unique][.filtered].predicted-orfs.bed.gz has a final output combining all predictions across your samples and/or replicates (including Bayes factor mean and variance for each sample, etc.). If you need a unique list of Ribo-seq ORFs (only coordinates i.e standard BED12, without duplicated entries for samples and/or replicates), use <project_name>[.note][-unique][.filtered].igv-orfs.bed.gz.
How to launch the web applications#
To launch the profile construction dashboard
rpbp-profile-construction-dashboard -c CONFIG
The application has multiple views to facilitate quality control, e.g.
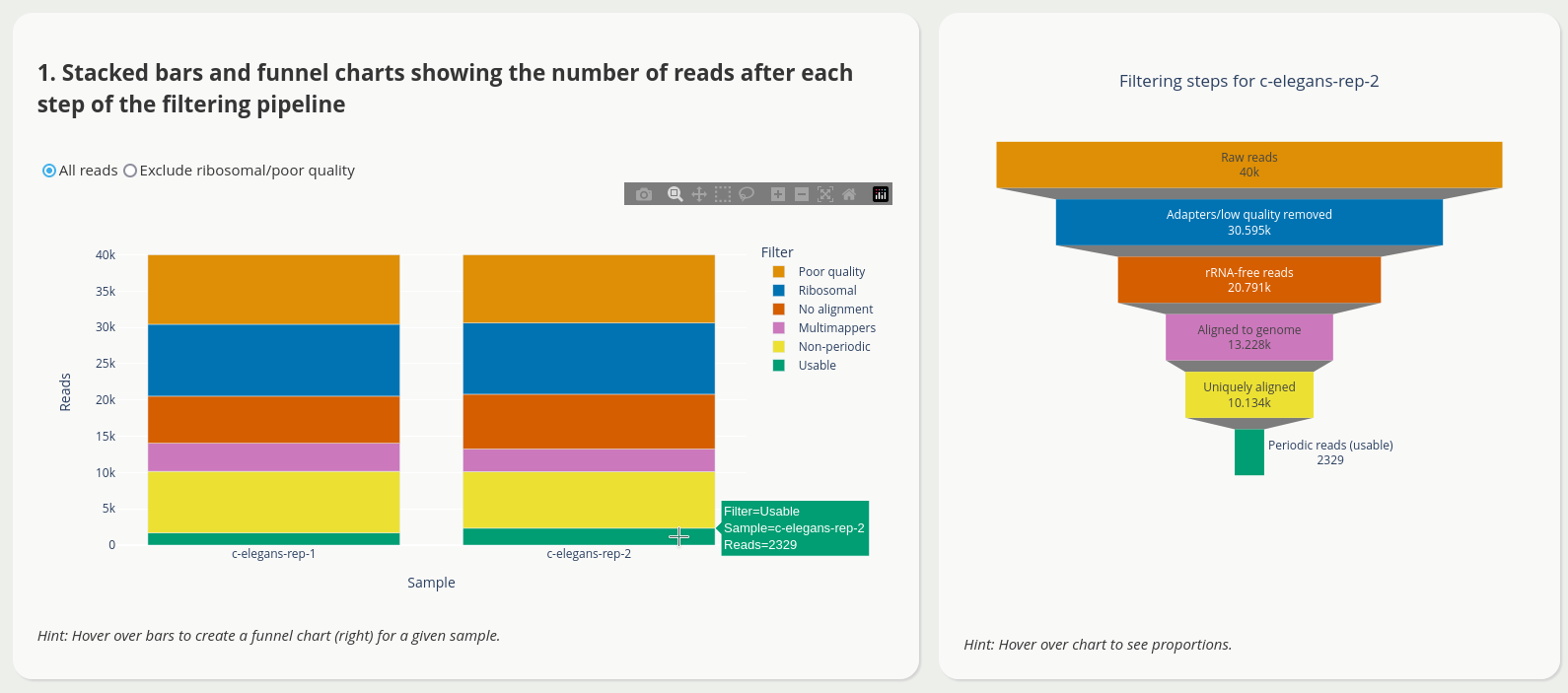
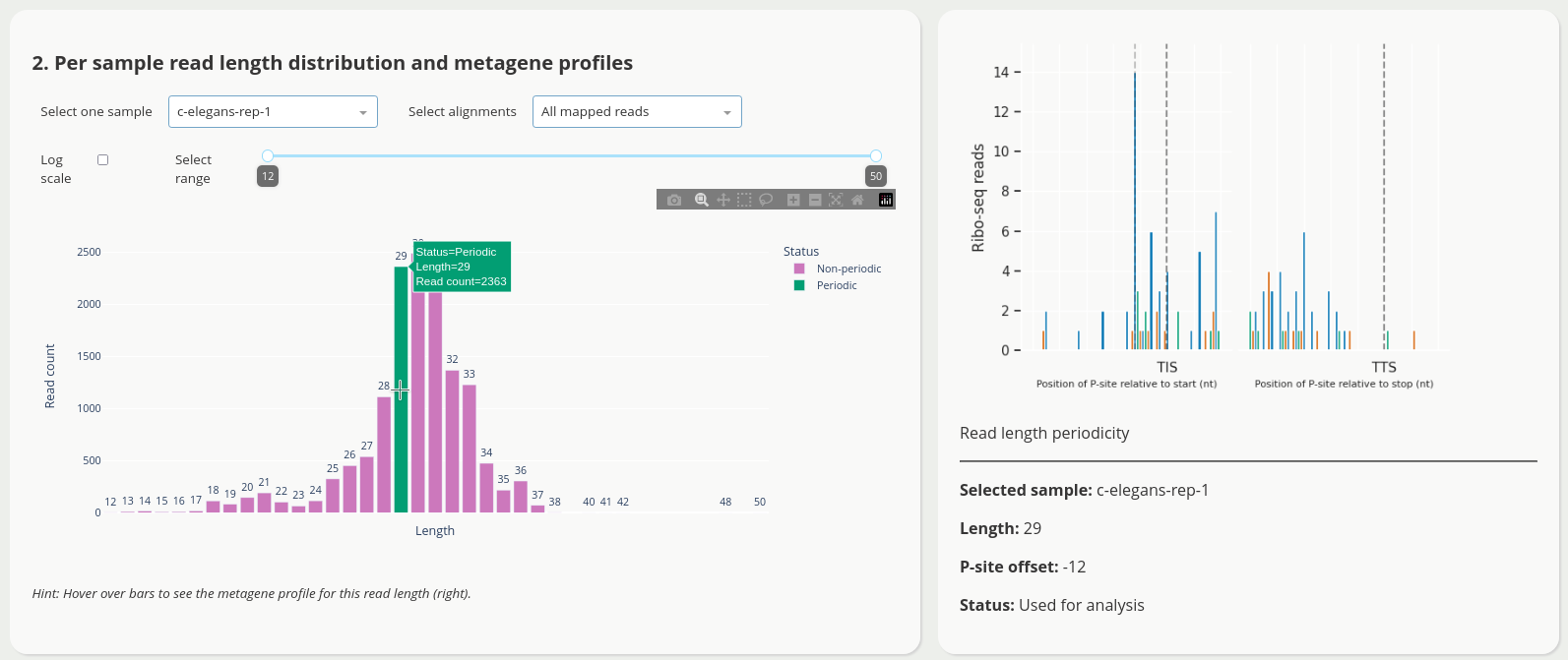
To launch the predictions dashboard
rpbp-predictions-dashboard -c CONFIG
The application has multiple views to facilitate ORF discovery, including an integrated IGV browser for the visual exploration of predicted Ribo-seq ORFs, e.g.
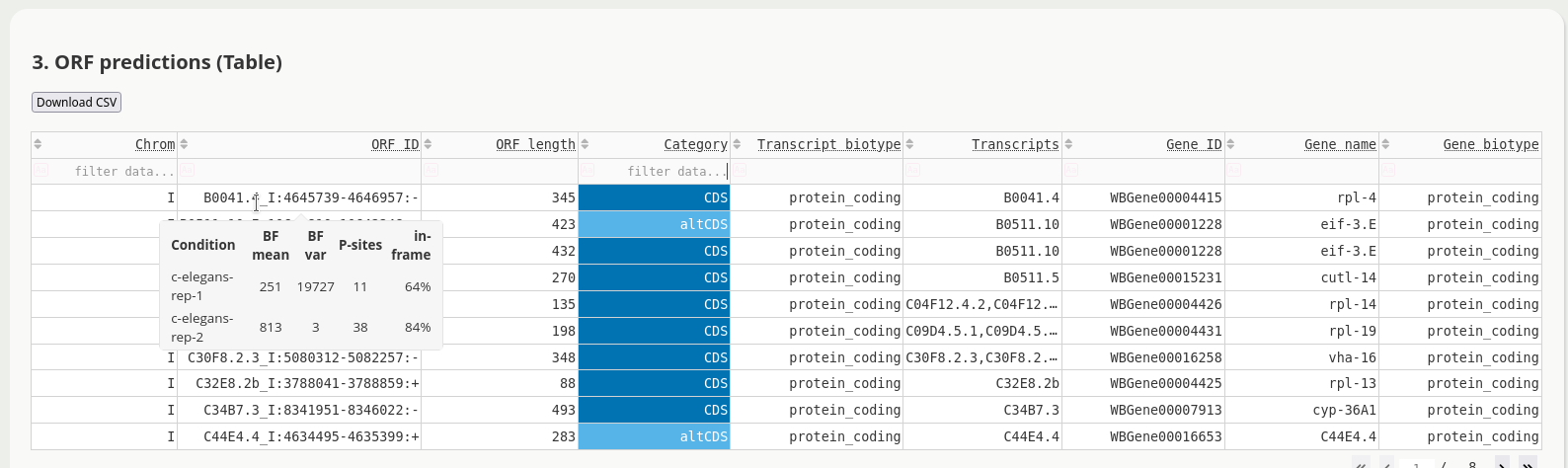
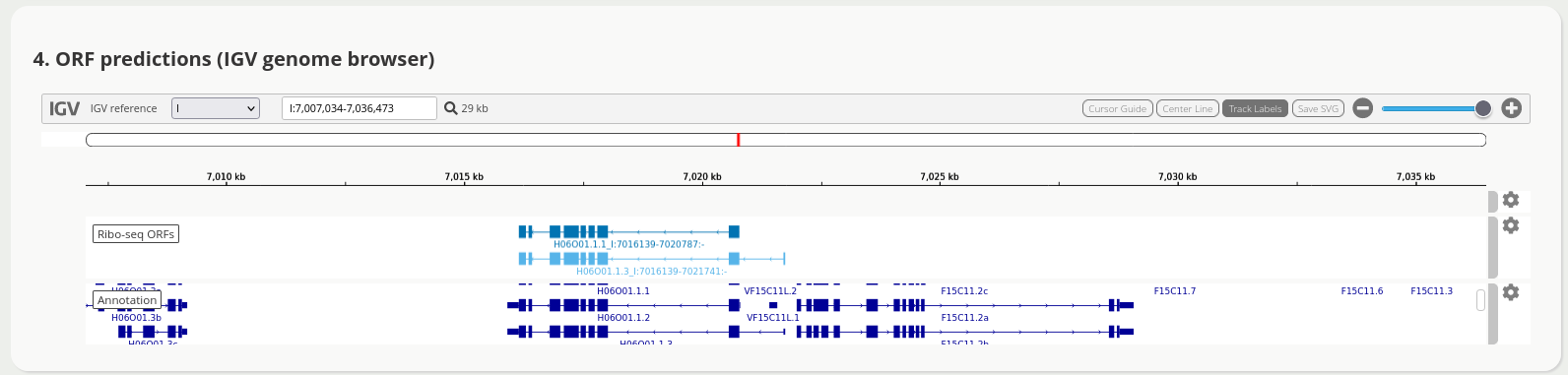
Try it out, and see more!
For all options, consult the API.
Note
Any of the above command will open a browser page with the web application running locally. You can also specify a --host and a --port, e.g. if launching the app from a remote server. In the latter case, you have to open a browser page at the correct address. For example, you use --host 123.123.123.123, then open a page on http://123.123.123.123:8050/.